8 August 2023
Antibiotic ribosomal compounds decipheredCombating Multidrug-Resistant Bacteria through High-Resolution Structural Imaging
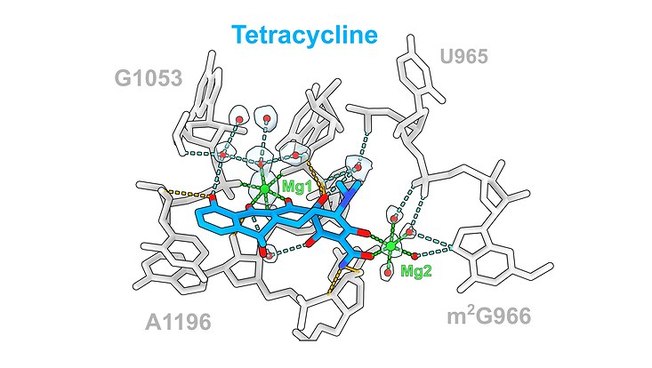
Photo: Universität Hamburg and Paternoga
Ribosomes are large molecular complexes found in plant, animal, human, and bacterial cells that produce proteins. They are important targets for antibiotics, and, in bacteria, the antibiotics bind to a subunit of the ribosomes and cause reading errors or even reading stops. This results in the formation of defective proteins that lose their biological function, and the bacteria then die. However, an increasing number of multidrug-resistant bacteria can prevent antibiotics from working.
A detailed understanding of antibiotics, in particular the antibiotic-ribosome structure, is essential to circumvent this resistance. Over the past two decades, antibiotic-ribosome structures have been published for each major class of ribosome-directed antibiotics, providing insights into their binding sites and mechanisms of action.
The team led by Prof. Dr. Daniel N. Wilson from the Department of Chemistry at Universität Hamburg has used cryoelectron microscopy to image these structures at a resolution of 1.6 to 2.2 angstroms (Å), where one Å is one ten-millionth of a millimeter. “In the past, antibiotic-ribosome structures have been imaged primarily at resolutions of 2.5 to 3.5 Å,” says Prof. Dr. Daniel N. Wilson. “The improved resolution even allows us to observe water molecules that support interactions between the drugs and the ribosomal components.” The researchers found that, in general, 10 to 20 hydrogen bonds interacted with the ribosome and that the level of interaction of the water molecules varied for the different classes of antibiotics.
The antibiotics studied included the 6 clinically relevant antibiotic classes, including the tetracyclines (respiratory infections), the broad-spectrum aminoglycosides (tuberculosis and meningitis), and the pleuromutilins (skin and soft tissue infections).
For the study, the research team applied these antibiotics to the 70S ribosomes of the bacterium Escherichia coli—a coliform bacterium that is also found in the human intestine. “We expect that this information can be used in the future for the structure-based design of new antibiotic derivatives. By identifying regions that can be altered, the bound water can be selectively displaced to interact with the target, which is the bacterium,” says Wilson.
High-resolution imaging is a big step forward, according to Wilson: “The old antibiotic-ribosome structures at lower resolutions show that the binding sites for each class of antibiotics are similar; in many cases, however, there are profound differences in terms of the exact position of the drugs as well as the drug binding site.” Wilson pointed out that the high-resolution experimental data were therefore needed to provide a more accurate description of the interactions of antibiotics with the ribosome.
In addition to Universität Hamburg, the study involved researchers from the Max Planck Institute for Multidisciplinary Natural Sciences in Göttingen, the Dubochet Center for Imaging at EPFL-UNIL in Switzerland, and the Central European Institute of Technology in the Czech Republic.
Original publication:
Paternoga, Crowe-McAuliffe, Bock, Koller, Morici, Beckert, Myasnikov, Grubmüller, Nováček, Wilson (2023) Structural conservation of antibiotic interaction with ribosomes, Nature Structural & Molecular Biology. https://doi.org/10.1038/s41594-023-01047-y
